Sitemap
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Pages
Welcome to my portfolio!
About me
Posts
Future Blog Post
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Blog Post number 4
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 3
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 2
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Blog Post number 1
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
portfolio
Multi-Omics Data Integration Using Machine Learning and Large Language Models (LLMs)
Duration: January 01, 2023 - October 01, 2023
Aims
- Integrate existing carotid plaque scRNAseq datasets with the current bestperforming integration methods.
- Benchmark these integration techniques and select the most suitable method for building a carotid plaque single-cell atlas.
- Identify subtypes of VSMCs and RNA markers for them.
- Study the abundances of the corresponding protein biomarkers in provided carotid plaque proteomics data and associate them with patient subgroups and plaque characteristics
- Return offer (internship) -> validating findings with bulk data, spatial data, pseudotime and cell-cell communication
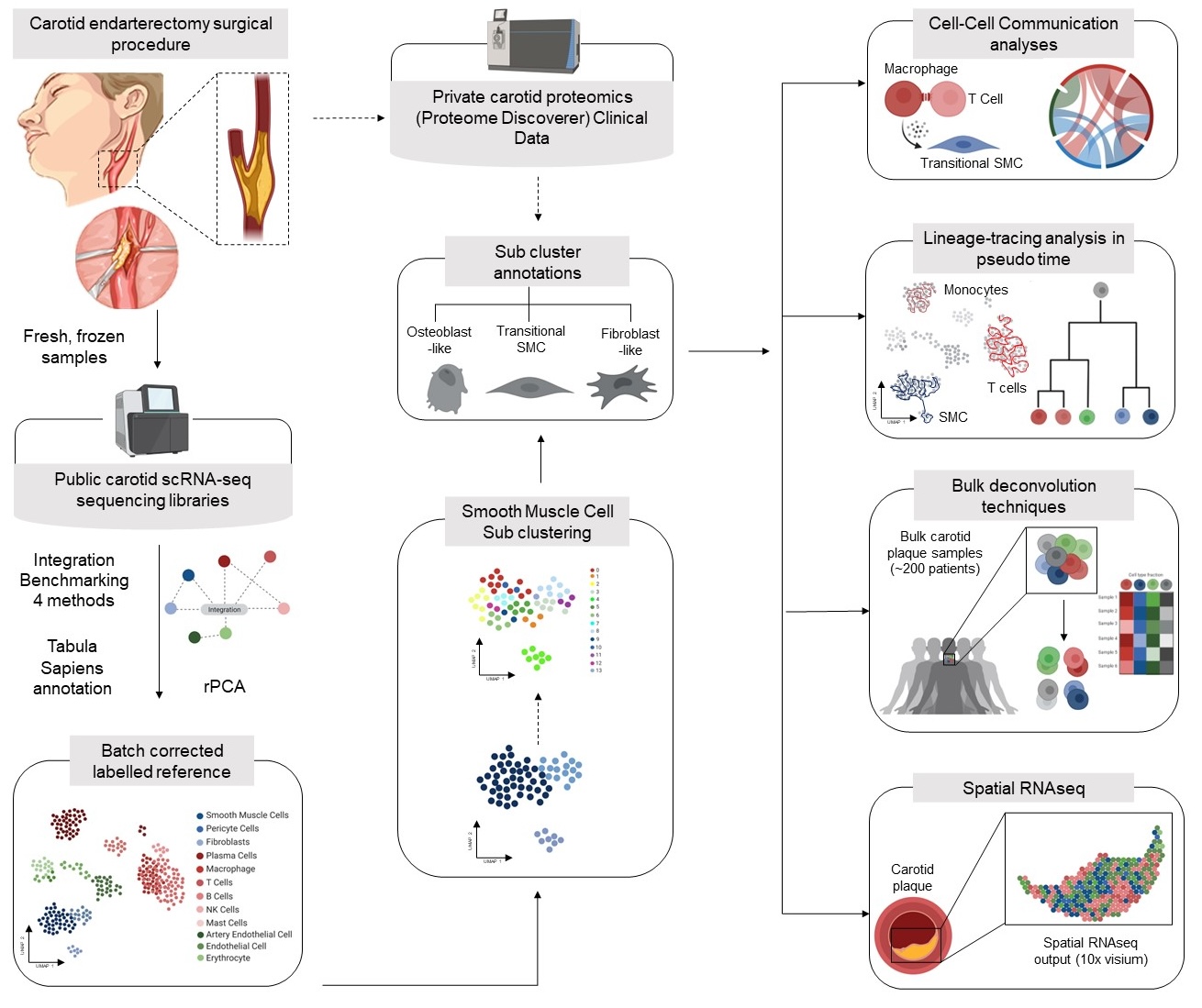
Figure 3: General Workflow of the project. Please click the title for more detailed information on results and outcomes.
Skills and Tools
| Task | Packages/Tools |
|---|---|
| Data Processing and Integration | |
| Single-cell RNA Sequencing |
|
| Multi-omics Integration |
|
| Proteomics |
|
Elucidating Spatial Cell Composition of Neuroblastoma (Group Project)
Duration: January 01, 2024 - March 01, 2024
Aims
- Meta-analyse various single-cell studies of neuroblastoma, building a single-cell atlas that spans tumour cell diversity (Figure 2.1)
- Identify cell populations and tissue heterogeneity in single cell spatial transcriptomics (SCST) data using transfer learning from the single-cell reference.
- Validate, test and scrutinise available tools for exploratory analysis of novel SCST data, including fine tuning and adpatation of a novel, multi-modal deep learning clustering approach (SiGra) (Figure 2.2)
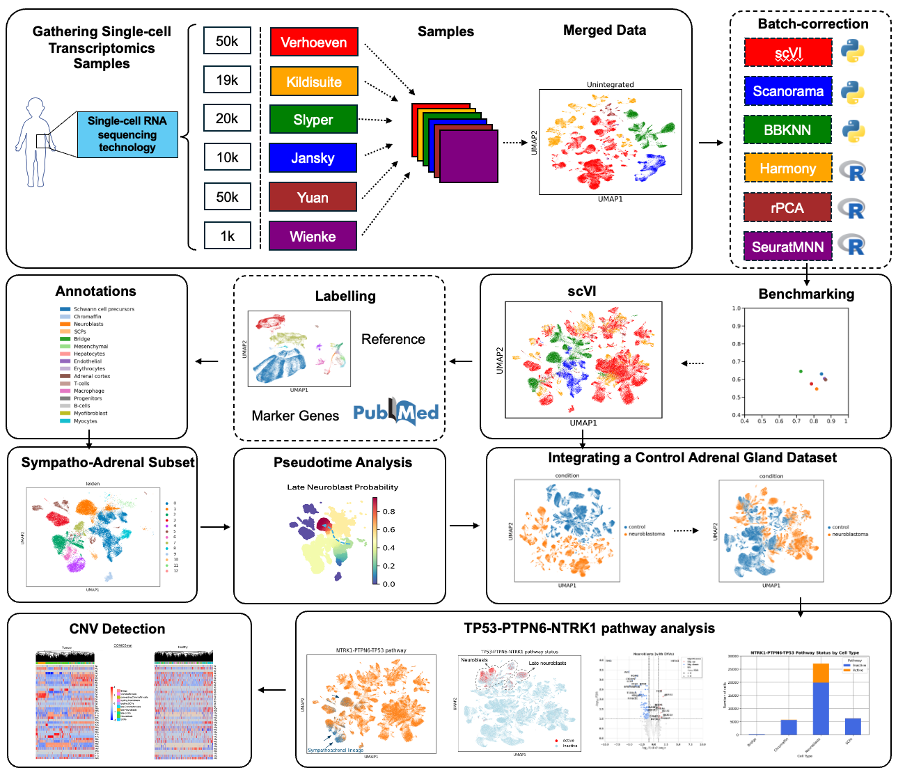
Figure 2.1: Initial, Single-cell RNA-Seq Workflow of the project. 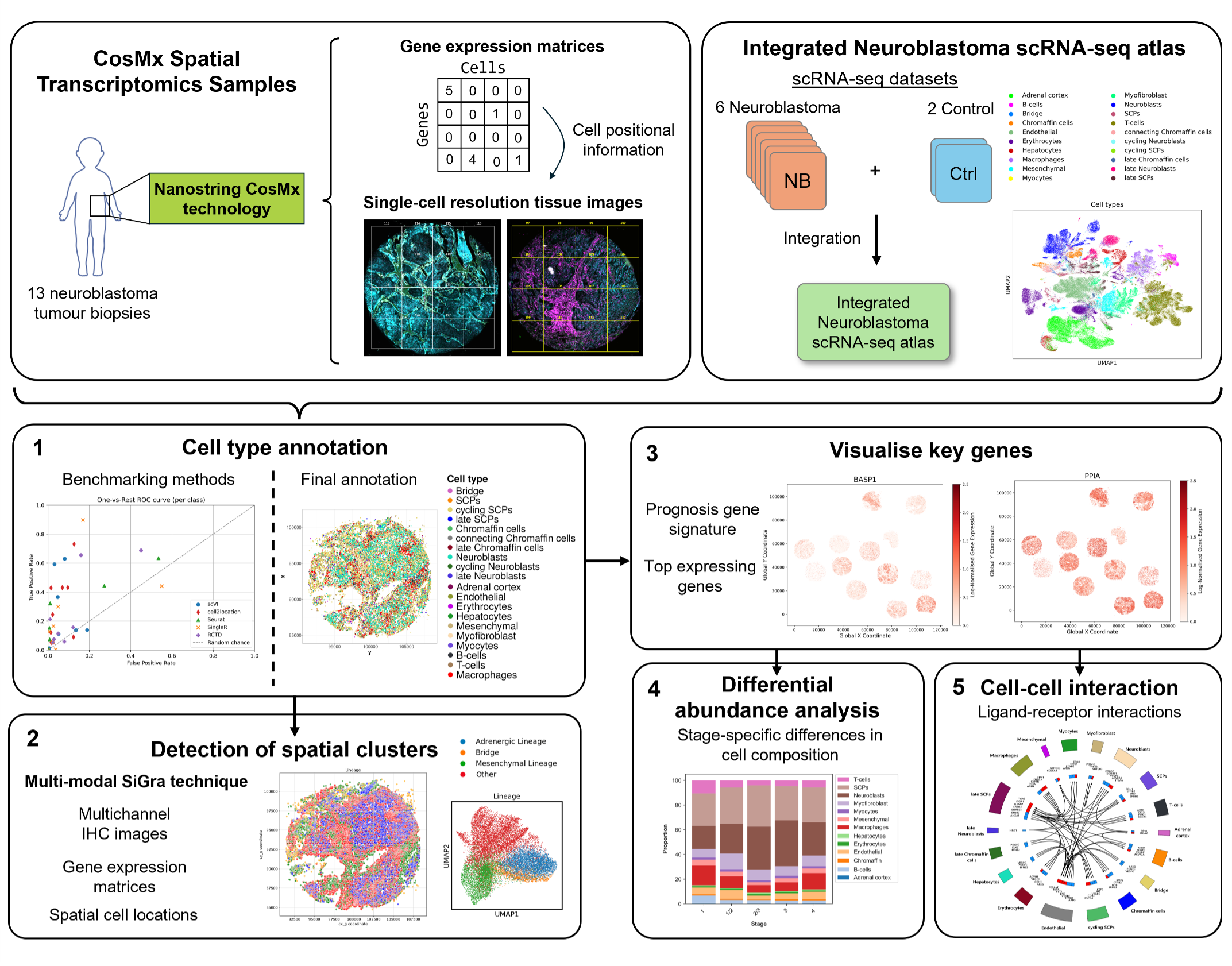
Figure 2.2: Secondary, Single-cell Spatial RNA-seq Workflow of the project. Please click the title for more detailed information on results and outcomes.
Skills and Tools
| Task | Packages/Tools |
|---|---|
| Single-Cell RNA-Seq Data Integration + Analysis |
|
| Single-Cell Spatial RNA-Seq Data Analysis |
|
| Deep Learning (SiGRA Modification) |
|
| Shell Computing |
|
Regulation and Biological Functions of Alternative Spilcing in Neurones of the Adult Mouse Visual Cortex
Duration: June 01, 2022 - September 01, 2022
Aims
- Process 4 single cell/single nuclear datasets from the Allen Brain Institute in R
- Generate high throughput splicing data using specialised tools in Unix (e.g Whippet).
- Identify evidence to support the mechanism and presence of NMD containing transcripts in the nuclei of PV interneurons
- Map different isoform profiles of GABAergic and glutamatergic layers
- Validate the utility of Single-Cell data for splicing analysis compared to Bulk RNA-seq
Figure 4: Poster displaying the outcome of this project investigating alternative splicing during Neurogenesis. Please click the title for more detailed information on results and outcomes.
Skills and Tools
| Task | Packages/Tools |
|---|---|
| Data Analysis |
|
| Single-cell RNA Sequencing |
|
| Multi-omics Integration |
|
| Proteomics |
|
Multi-Omics Pathway-based Data Integration Using Machine Learning and Large Language Models (LLMs)
Duration: March 01, 2024 - October 01, 2024
Aims
- Use LLM-based semantic searching to map more metabolite names to IDs, widening the scope of pathway analysis (Figure 1A)
- Apply novel pathway databases augmented with metabolites using deep learning, facilitating more comprehensive pathway mapping and attaining more robust biological predictions (Figure 1B)
- Develop an extension to PathIntegrate which will provide an unsupervised utility for pathway-based multivariate analysis that can be benchmarked with synthetic data simulations (Figure 1C)
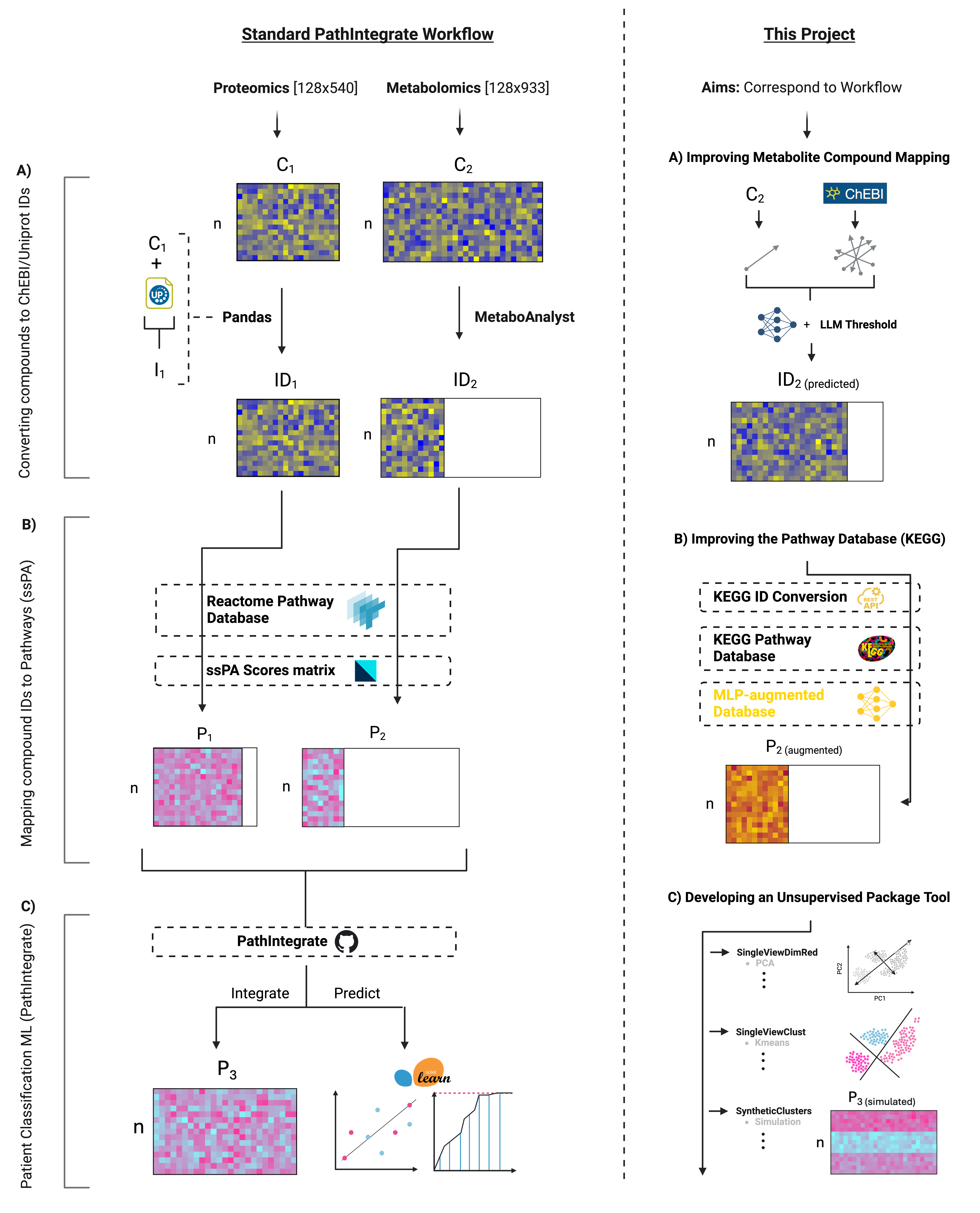
Figure 1: General Workflow of the project. Please click the title for more detailed information on results and outcomes.
Skills and Tools
| Task | Packages/Tools |
|---|---|
| Data Processing, Analysis and Databases |
|
| Machine Learning + Web App |
|
| Miscellaneous |
|
| Bioinformatics Tools |
|
publications
Alternative Splicing and Neurogenesis
Published in KCL Biochemist Print, 2021
This article was inspired by the work of Professor Eugene Makeyev who studied intricate processes of gene expression regulation, such as NMD, during neurogenesis. My interest in this biological problem inspired my first bioinformatics internship which kickstarted my journey in the field today
The Science and Economics of Cannabis: A Case Study of Thailand
Published in Kings Business Review, 2022
This article is about the economic impact of cannabis in Thailand, also considering the scientific effects of cannabis on the brain and body. Please click the title if you want to have a read.
Novel Plastic Degrading Enzymes
Published in KCL Biochemist Print, 2022
This article was inspired by the work of Professor Zelesniak who studied plastic degrading enzymes. It covers key processes of bioremediation and the potential of these enzymes for efficient recycling.
Proteomic Atlas of Atherosclerosis: The Contribution of Proteoglycans to Sex Differences, Plaque Phenotypes, and Outcomes
Published in Circulation Research, 2023
I was a co-author for this publication, working on the single-cell component. This study provides a comprehensive proteomics analysis of human atherosclerotic plaques, revealing sex differences and distinct plaque phenotypes with prognostic implications.
GitHub Repository: https://github.com/Cardiovascular-Bioinformatics/Plaques_Cellular_Composition
Citation: Theofilatos K, Stojkovic S, Hasman M, Popham J, et al. (2023). "Proteomic Atlas of Atherosclerosis: The Contribution of Proteoglycans to Sex Differences, Plaque Phenotypes, and Outcomes." Circulation Research. 133(7):542-558. doi:10.1161/CIRCRESAHA.123.322590.
Visit Paper
MarketsAndMarkets 2nd Annual Single Cell-Omics Conference
Invited to the MarketsAndMarkets Conference, 2023
This conference discussed various Single-Cell RNA-Seq platforms available and emerging Single-Cell research in the field.
PathIntegrate: Multivariate Modelling Approaches for Pathway-Based Multi-Omics Data Integration
Published in PLOS Computational Biology, 2024
I contributed to the development of the PyPi PathIntegrate Python package (included in v1.0.0), a tool for pathway-based multi-omics data integration used in this publication. My contribution has a seperate doi but it is based on this paper from PLOS.
GitHub Repository: https://github.com/cwieder/PathIntegrate
Citation: Popham, J., Wieder, C., & Ebbels, T. (2024). cwieder/PathIntegrate: PathIntegrate Unsupervised (v1.0.0). Zenodo. --> 'Wieder C, Cooke J, Frainay C, et al. (2024). PathIntegrate: Multivariate Modelling Approaches for Pathway-Based Multi-Omics Data Integration. PLOS Computational Biology. 20(3): e1011814. doi:10.1371/journal.pcbi.1011814.
Visit Paper
Spatial Biology Network KCL
Invited to the Kings Spatial Biology Research Conference, 2024
This conference was focused on pilot studied with revolutionary CosMX Nanostring Spatial RNA-Seq data. All research groups, including ours, were working within the KCL Spatial Biology Network. Click the title to see an image of our presentation.
talks
Talk 1 on Relevant Topic in Your Field
Published:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Conference Proceeding talk 3 on Relevant Topic in Your Field
Published:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.
teaching
Raw RNA-Seq Data Analysis Project (R and Unix)
For this project, I conducted a comprehensive RNA-seq data analysis to explore gene expression differences between experimental groups. First, I carried out quality control checks on the raw sequencing data to assess read quality, using tools such as Rsubread to extract quality scores and visualize them with boxplots. This step ensured that the data was of sufficient quality for reliable downstream analysis.
Sequence Generator Project (Python)
I developed a Python program to generate synthetic promoter sequences with specific characteristics to simulate and study promoter regulation. The project aimed to help in understanding how different motifs within a promoter region influence transcription.
