Multi-Omics Pathway-based Data Integration Using Machine Learning and Large Language Models (LLMs)
Duration: March 01, 2024 - October 01, 2024
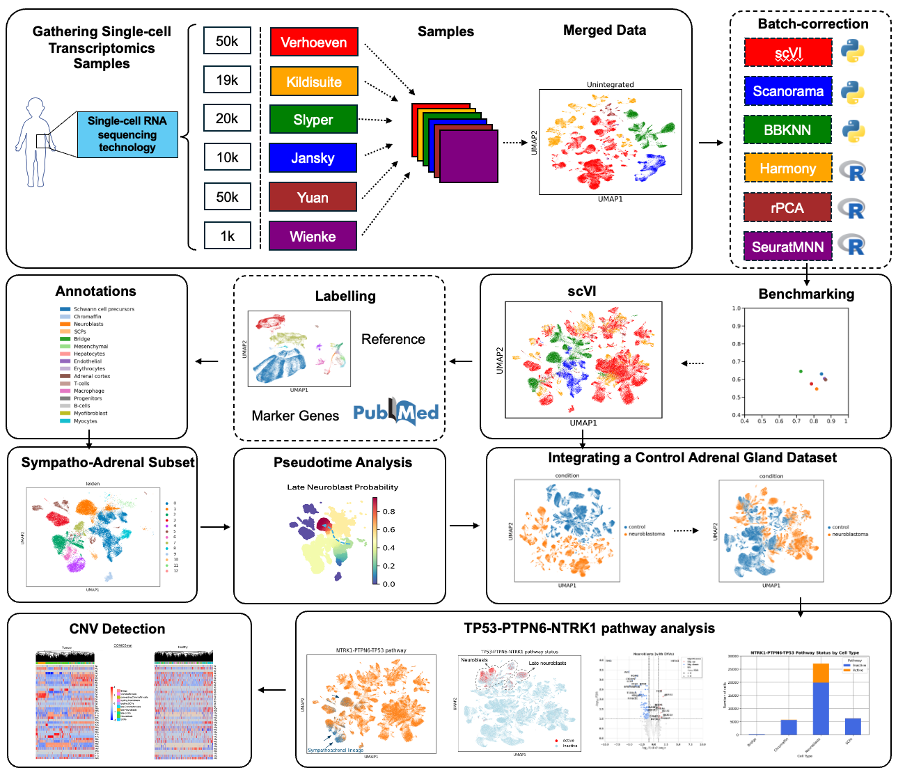
Aims
- Use LLM-based semantic searching to map more metabolite names to IDs, widening the scope of pathway analysis (Figure 1A)
- Apply novel pathway databases augmented with metabolites using deep learning, facilitating more comprehensive pathway mapping and attaining more robust biological predictions (Figure 1B)
- Develop an extension to PathIntegrate which will provide an unsupervised utility for pathway-based multivariate analysis that can be benchmarked with synthetic data simulations (Figure 1C)
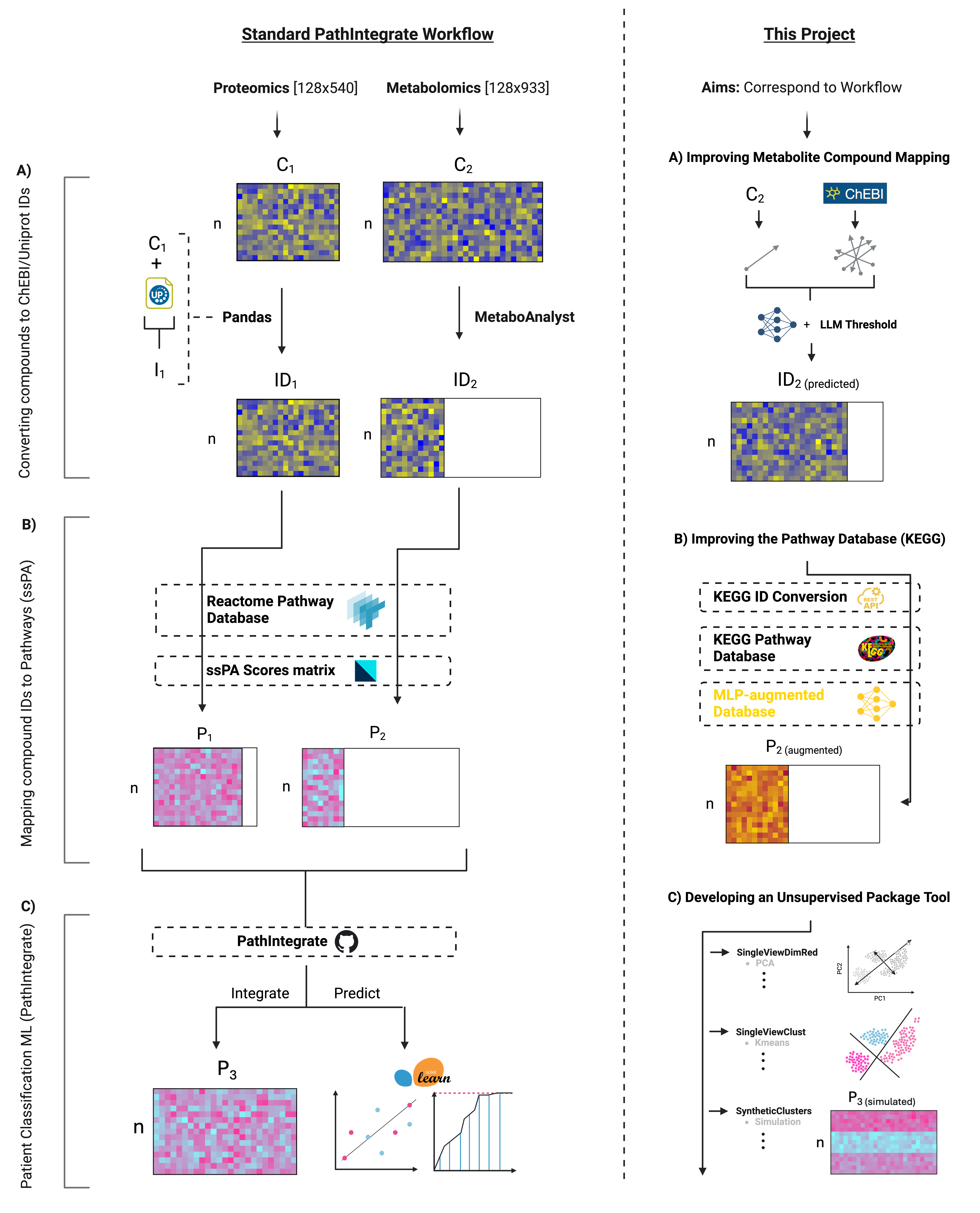
Figure 1: General Workflow of the project. Please click the title for more detailed information on results and outcomes.
Skills and Tools
| Task | Packages/Tools |
|---|---|
| Data Processing, Analysis and Databases |
|
| Machine Learning + Web App |
|
| Miscellaneous |
|
| Bioinformatics Tools |
|